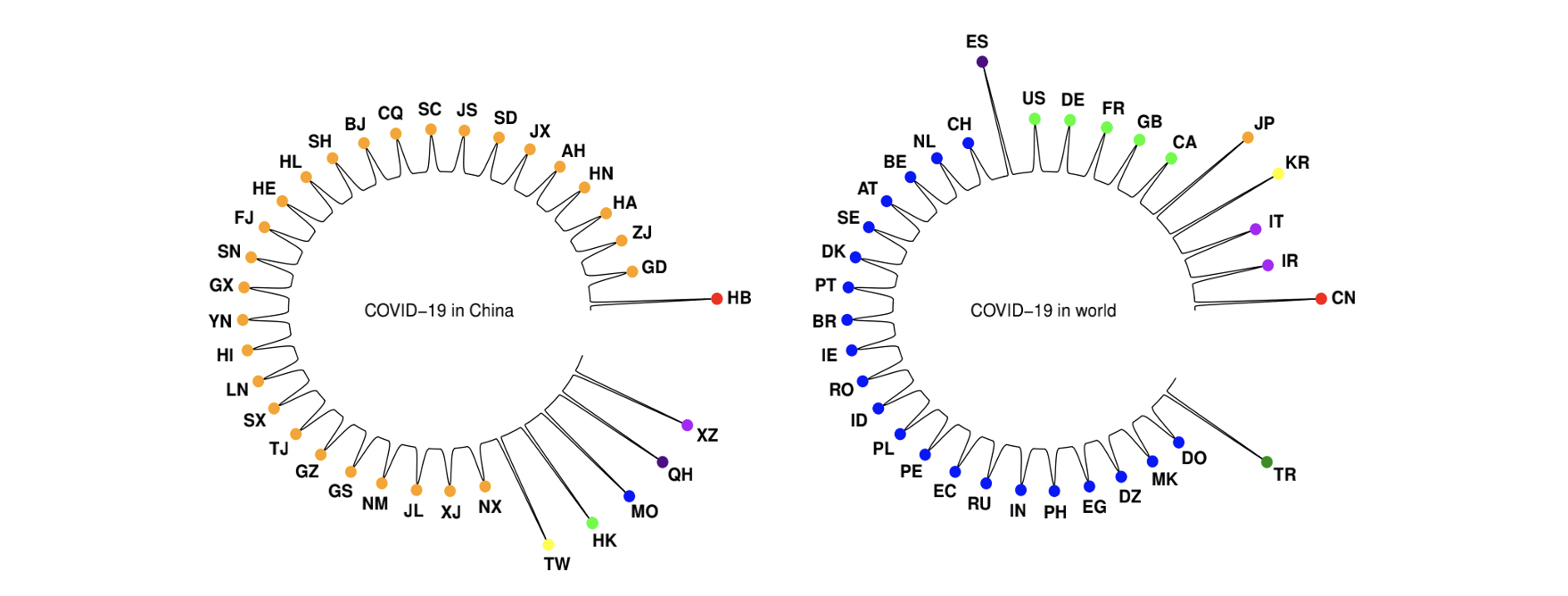
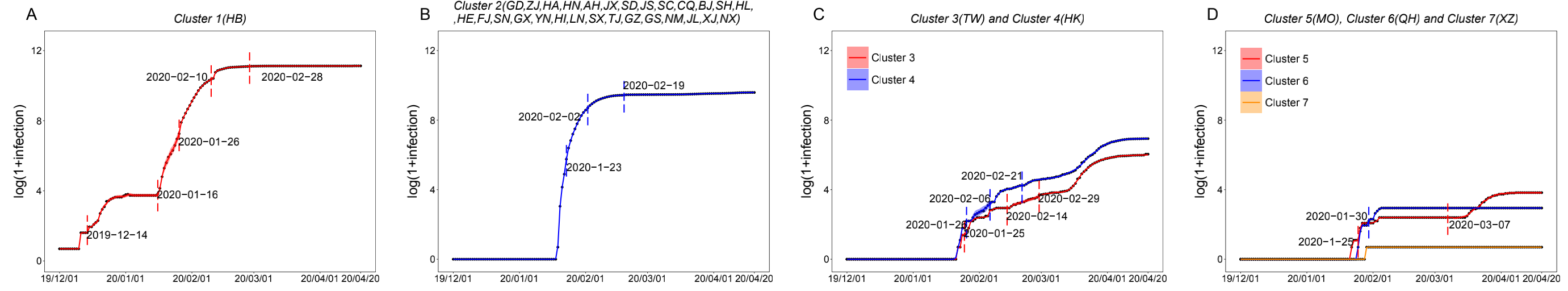
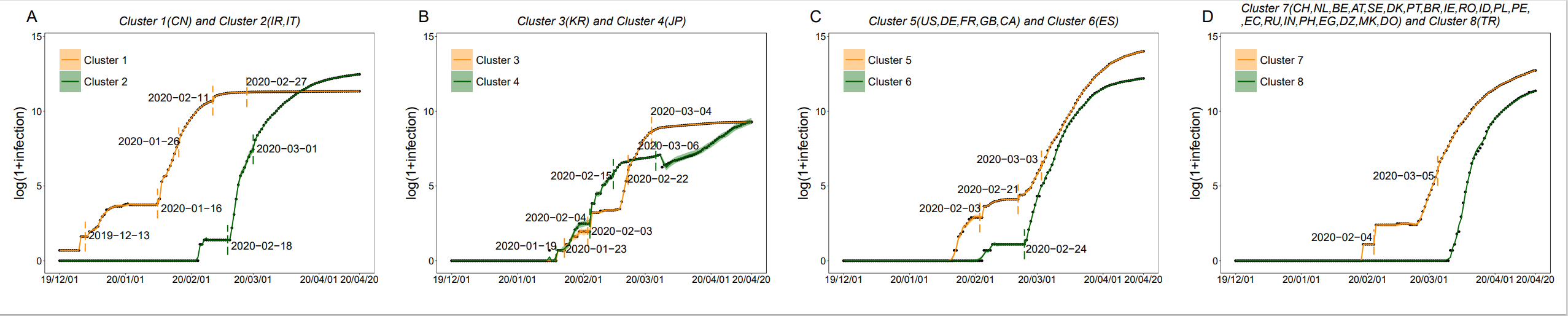
Graph Clustering System (GCS)
If there are some problems with the page display, you can change your browser and try it (Google Chrome is recommended).
If you are interested in GCS, please be sure to read the supporting document "Supplemental document".